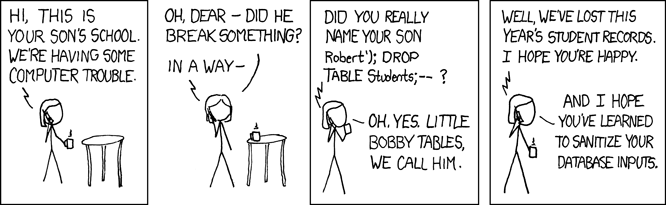
Lesson 5 Working with relational data & databases
5.1 Relational data in tidyverse
We spend quite some time now on learning how to visualise, analyse, and describe data. But where do we get the data? In this lesson you will learn how to interact with databases using the SQL language.
There is quite a lot of material in this lesson, but the exercises within the lesson are short. You won’t be an absolute SQL wizard after this afternoon. We are aiming on giving you enough of a steady start with DBeaver and SQL to build on when you may encounter working with SQL in the future. But at times it will feel pretty cool, we promise!
5.1.1 Relational data
It is quite rare for a data analysis to use only one table of data. Suppose you investigate the effects of a certain drug on behaviour of rats in a maze. You may have blood levels of the drug at certain times, some automatic recording of movements, maybe you weighted them before every experiment, etc. You may enter some of these values in excel, but other values about the same rats will be stored in different formats or in different files, such as the video recordings or descriptive statistics of these video recordings (average time in maze, number of mistakes etc etc). Still, you will want to look at all this information combined. Collectively, these tables of data are called relational data.
For example, we used the NYflights13 package before to look at relational data in DAUR1. It contains several tables with information, which are related. In the diagram below, which we borrowed from r4ds by Hadley Wickham and Garrett Grolemund, some of the variables in the different tables are depicted:
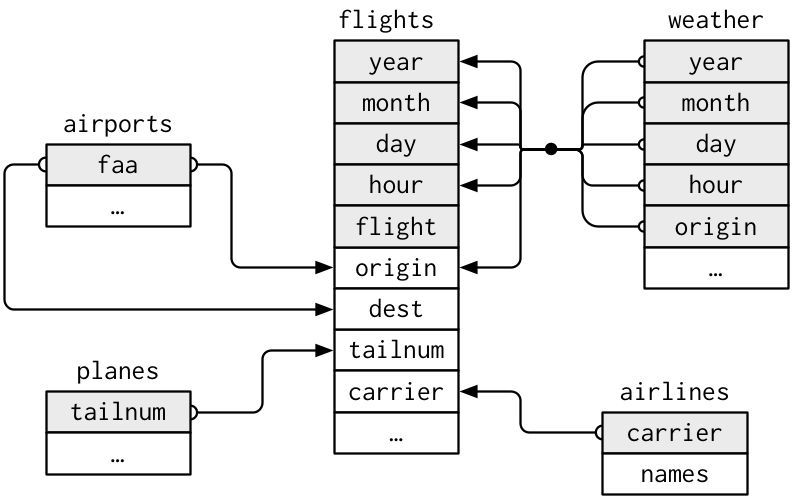
You can see that some variables in one table are present or translate to variables in other tables. Therefore, the tables can be combined. If you want more information on the plane used on a flight in the flights table, you can use it’s tailnumber to look it up in the planes table (or actually, have R do it for you by joining the tables, don’t look it up manually obviously…)
5.1.2 Remember tidy data?
We will start this lesson with a rehearsal of things you did in DAUR1 and 2.
The nyflights tables are really nice and tidy. You have used them before to practice joining tibbles, and they are joined quite easily.
Quite often if you download or gather data and want to join two datasets, you may need to reshape them. Usually, having or transforming your data in the tidy shape is a good practice if you want to perform subsequent joins.
Tidy data means:
- All variables in their own column
- All observations in their own row
- Every cell contains a single observations
You may have used gather() and spread() in DAUR1 to make tibbles tidy or untidy depending on your needs. They have been updated to be a bit more user friendly! The new functions are called pivot_longer and pivot_wider:
Here is an example with pivot_longer() :
library(palmerpenguins)
library(tidyverse)
penguins %>%
tidyr::pivot_longer(contains("_"), # measurement cols
names_to = c("variable_name"),
values_to = "value")## # A tibble: 1,376 × 6
## species island sex year variable_name value
## <fct> <fct> <fct> <int> <chr> <dbl>
## 1 Adelie Torgersen male 2007 bill_length_mm 39.1
## 2 Adelie Torgersen male 2007 bill_depth_mm 18.7
## 3 Adelie Torgersen male 2007 flipper_length_mm 181
## 4 Adelie Torgersen male 2007 body_mass_g 3750
## 5 Adelie Torgersen female 2007 bill_length_mm 39.5
## 6 Adelie Torgersen female 2007 bill_depth_mm 17.4
## 7 Adelie Torgersen female 2007 flipper_length_mm 186
## 8 Adelie Torgersen female 2007 body_mass_g 3800
## 9 Adelie Torgersen female 2007 bill_length_mm 40.3
## 10 Adelie Torgersen female 2007 bill_depth_mm 18
## # ℹ 1,366 more rowsOr perhaps (see the difference?):
penguins_long <- penguins %>%
tidyr::pivot_longer(contains("_"), # measurement cols
names_to = c("part", "measure", "unit"),
names_sep = "_")
penguins_long## # A tibble: 1,376 × 8
## species island sex year part measure unit value
## <fct> <fct> <fct> <int> <chr> <chr> <chr> <dbl>
## 1 Adelie Torgersen male 2007 bill length mm 39.1
## 2 Adelie Torgersen male 2007 bill depth mm 18.7
## 3 Adelie Torgersen male 2007 flipper length mm 181
## 4 Adelie Torgersen male 2007 body mass g 3750
## 5 Adelie Torgersen female 2007 bill length mm 39.5
## 6 Adelie Torgersen female 2007 bill depth mm 17.4
## 7 Adelie Torgersen female 2007 flipper length mm 186
## 8 Adelie Torgersen female 2007 body mass g 3800
## 9 Adelie Torgersen female 2007 bill length mm 40.3
## 10 Adelie Torgersen female 2007 bill depth mm 18
## # ℹ 1,366 more rowsExercise 5
What is the first variable in the penguins_long dataframe printed above?
Click for the answer
species (just to make sure you still know which parts of a tibble/dataframe are variables)Let’s get it back to wide again:
penguins_long %>%
tidyr::pivot_wider(names_from = c("part", "measure", "unit"), # pivot these columns
values_from = "value", # take the values from here
names_sep = "_") # combine col names using an underscore## Warning: Values from `value` are not uniquely identified; output will contain list-cols.
## • Use `values_fn = list` to suppress this warning.
## • Use `values_fn = {summary_fun}` to summarise duplicates.
## • Use the following dplyr code to identify duplicates.
## {data} |>
## dplyr::summarise(n = dplyr::n(), .by = c(species, island, sex, year, part,
## measure, unit)) |>
## dplyr::filter(n > 1L)## # A tibble: 35 × 8
## species island sex year bill_length_mm bill_depth_mm flipper_length_mm
## <fct> <fct> <fct> <int> <list> <list> <list>
## 1 Adelie Torgersen male 2007 <dbl [7]> <dbl [7]> <dbl [7]>
## 2 Adelie Torgersen female 2007 <dbl [8]> <dbl [8]> <dbl [8]>
## 3 Adelie Torgersen <NA> 2007 <dbl [5]> <dbl [5]> <dbl [5]>
## 4 Adelie Biscoe female 2007 <dbl [5]> <dbl [5]> <dbl [5]>
## 5 Adelie Biscoe male 2007 <dbl [5]> <dbl [5]> <dbl [5]>
## 6 Adelie Dream female 2007 <dbl [9]> <dbl [9]> <dbl [9]>
## 7 Adelie Dream male 2007 <dbl [10]> <dbl [10]> <dbl [10]>
## 8 Adelie Dream <NA> 2007 <dbl [1]> <dbl [1]> <dbl [1]>
## 9 Adelie Biscoe female 2008 <dbl [9]> <dbl [9]> <dbl [9]>
## 10 Adelie Biscoe male 2008 <dbl [9]> <dbl [9]> <dbl [9]>
## # ℹ 25 more rows
## # ℹ 1 more variable: body_mass_g <list>Oh, we’re in trouble. Read the warning message.
Exercise 5
What happened? Can you think of a way we could have prevented this error a couple of steps before? Try it!
Click for the answer
R isn’t sure where to put the data anymore, as there are duplicates. Every measurement was not marked by some unique code and there are (for instance) 7 male Adelie penguin measured on Torgersen in 2007, and 5 female Adelie penguin measured on Biscoe in 2007. Now what?
We need to be able to identify each measurement from the other ones based on something else.
For instance, just number all the measurements in the first place: (note that this is just an example of a solution, anything you came up with that works, works!)
# give each measured penguin a number
penguins_with_id <- penguins %>% mutate(id=seq(nrow(.)))
# make the data long again
penguins_long_ided <- penguins_with_id %>%
tidyr::pivot_longer(contains("_"), # measurement cols
names_to = c("part", "measure", "unit"),
names_sep = "_")
# and wide again
penguins_long_ided %>%
tidyr::pivot_wider(names_from = c("part", "measure", "unit"),
values_from = "value",
names_sep = "_") ## # A tibble: 344 × 9
## species island sex year id bill_length_mm bill_depth_mm
## <fct> <fct> <fct> <int> <int> <dbl> <dbl>
## 1 Adelie Torgersen male 2007 1 39.1 18.7
## 2 Adelie Torgersen female 2007 2 39.5 17.4
## 3 Adelie Torgersen female 2007 3 40.3 18
## 4 Adelie Torgersen <NA> 2007 4 NA NA
## 5 Adelie Torgersen female 2007 5 36.7 19.3
## 6 Adelie Torgersen male 2007 6 39.3 20.6
## 7 Adelie Torgersen female 2007 7 38.9 17.8
## 8 Adelie Torgersen male 2007 8 39.2 19.6
## 9 Adelie Torgersen <NA> 2007 9 34.1 18.1
## 10 Adelie Torgersen <NA> 2007 10 42 20.2
## # ℹ 334 more rows
## # ℹ 2 more variables: flipper_length_mm <dbl>, body_mass_g <dbl>5.1.3 Joining data in databases
Joining operations are frequently used to increase or reduce the amount of data you have. To join tables you need some shared information to uniquely identify each observation: a key. In simple cases, one single variable is sufficient to identify each measurement (simple key). Each car in the Netherlands is identified by its license plate. And for example, our fictional rabbit ear length dataset could look like this, with a key variable in the first column:
| rabbit_id | time | oorlengte |
|---|---|---|
| rabbit_1 | 2015-11-24 | 8.076705 |
| rabbit_2 | 2015-12-15 | 9.767767 |
| rabbit_3 | 2016-01-05 | 6.602540 |
| rabbit_4 | 2016-01-26 | 8.600057 |
| rabbit_5 | 2016-02-16 | 6.654945 |
| rabbit_6 | 2016-03-08 | 6.801595 |
Let’s check if they are indeed unique:
## # A tibble: 0 × 2
## # ℹ 2 variables: rabbit_id <chr>, n <int>Exercise 5
Read the output. Why does this give an empty tibble? Is that a good thing?
Click for the answer
There are no rabbit_id’s that are used more than once in the dataset. That is exactly what we wanted.As you have seen in the previous paragraph, the penguins dataset in palmerpenguins does not have one key variable. There is no unique code to identify each measured penguin. If two separate penguins by some extreme coincidence have the exact same metrics (same year, same species, island, same bill length, same body mass, same everything), their rows would be identical.
## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <fct> <fct> <dbl> <dbl> <int> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## # ℹ 2 more variables: sex <fct>, year <int>You cannot identify an observation by using a single variable (“this penguins bill length is 41.1”).
But if you combine several columns, you can identify a penguin (compound key e.g. “this penguin has bill length 39.5, flipper length 186 and is female”). As you have seen, not just any combination of variables will work (species + island + sex + year was not enough: “this male Adelie penguin was measured on Torgersen in 2007” will yield you 7 possible penguins.).
Suppose you have some dataset and want to add a simple key, the easiest way is just to number your measurements as we did before in the example solution to one of the previous questions (I like my ID’s to be in the first few columns, so I use “.before=”) :
## # A tibble: 344 × 9
## penguin_id species island bill_length_mm bill_depth_mm flipper_length_mm
## <int> <fct> <fct> <dbl> <dbl> <int>
## 1 1 Adelie Torgersen 39.1 18.7 181
## 2 2 Adelie Torgersen 39.5 17.4 186
## 3 3 Adelie Torgersen 40.3 18 195
## 4 4 Adelie Torgersen NA NA NA
## 5 5 Adelie Torgersen 36.7 19.3 193
## 6 6 Adelie Torgersen 39.3 20.6 190
## 7 7 Adelie Torgersen 38.9 17.8 181
## 8 8 Adelie Torgersen 39.2 19.6 195
## 9 9 Adelie Torgersen 34.1 18.1 193
## 10 10 Adelie Torgersen 42 20.2 190
## # ℹ 334 more rows
## # ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>So to recap: In this example we made a key variable with a unique value for each penguin: a simple key. But when searching through a database, a key can also be a combination of attributes (columns): a compound key. As long as this combination lets you uniquely identify records (rows) in the table.
Keys can be used to join different tables. You have used this before in DAUR1.
Exercise 5
Let’s refresh your memory. Take the penguins dataset.
As we all know, Adelie penguins love peanut butter, Chinstrap penguins prefer cheese and Gentoo penguins do anything for chocolate.
Here is a table describing food preference:
food_preference <-
tibble(
penguin_species = c("Adelie", "Chinstrap", "Gentoo"),
favorite_food = c("peanut butter", "cheese", "chocolate")
)Join this table with the penguins table in such a way that favorite_food becomes a variable within penguins.
What is/are the key variables?
Click for the answer
The key variables are species and penguin_species. ‘species’ here can be called the “primary key” as it is the key variable in the table we are working with primarily. The counterpart key variable in another table (in this case ‘penguin_species’) is called the “foreign key”.Exercise 5
Now make a new tibble containing only the information on penguin species, sex and bill length measured in 2009 using filter and select.
Click for the answer
You should really revisit DAUR1, this lesson here in the link if you forgot how to do this.
Note: as you filter on year, but year is not part of the selection, filter first, select second.5.2 Databases in the wild
We spent some time in R to get familiar with the act of joining, filtering, playing with data again. And we introduced keys: “primary key” in the table we are working with primarily, and the counterpart key variable in another table: the “foreign key”.
Now suppose you want to access some other, perhaps company specific database, which does not have its neatly described bioconductor package or is included in an R package. The lingua franca for retrieving (and transform, manage and store) information from relational databases is Structured Query Language - or SQL . You can pronounce this “S-Q-L” (in English) or “sequel”, both are used. It was already developed in the 60’s and has grown into a very accessible and popular language (with a few dialects..) to communicate with databases.
Watch this GLITCH video on SQL, because it is very clear and uses LEGO! (seriously, watch it, it is only 4 minutes.)
5.2.1 Relational databases
We have discussed relational databases in DAUR2, but what were they again? In a relational database, data is organised in -mostly- tables that can be linked to each other based on data that is available in both tables (remember the left_join() function?). These tables are called relations, and columns are not called variables but attributes or fields, and rows are called tuples or records. Luckily, SQL just calls them rows, columns and tables. But make note of the terms because you may encounter them when diving in databases.
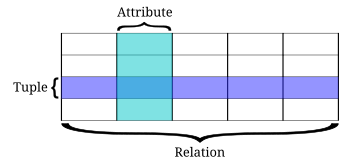
Figure 5.1: Relational databases and their preference for not just naming rows ‘rows’.
5.2.2 database management stystems
These relational databases can be stored on your computer or on a server you have direct access to, and managed in different ways. One popular system is PostgreSQL (often pronounced without the Q and L), used for instance Instagram, Spotify, Reddit and Netflix. It is not (yet) the most used system, according to a Stack Overflow Developer Survey (see eversql.com here), that would be MySQL. But as PostgreSQL is growing very fast, and actually works about the same as MySQL but with some additional possibilities, we will focus on PostgresQL. PostGreSQL is one possible way to store, retrieve and update (relational) data in a database.
We will discuss how to create databases, create or join tables within databases, how to insert records, delete or update them. Also we will do some data wrangling with SQL.
5.3 Introduction to DBeaver and SQL
In order to work with a database, we will need access to one. You can actually use SQL in the terminal, but we will use software instead: DBeaver. One of the prerequisites for this course was installing DBeaver. You may have clicked around in DBeaver a bit, which would be great. We will walk you through some steps, you are required to do them too. Not just read about it.
Open DBeaver.
Upon installation, DBeaver will have asked you if you wanted a sample database. If you didn’t, click help –> Create Sample Database. It will show up on the left part of the screen. This is not MySQL or PostgreSQL, but a SQlite database, but it will work just fine to have a look around.
Figure 5.2: Starting DBeaver for the first time
Double click on the sample database, and agree to download some drivers.
Now below the sample database item are several contents: tables, views, indexes etc.
Open Tables and double click on Album. This in one of the available tables in de database. You are presented with some meta data (columns, keys, foreign keys etc).
Exercise 5
To which table is the foreign key in this table linking?
Click for the answer
Artist
To see the actual data, you can click on the data tab:
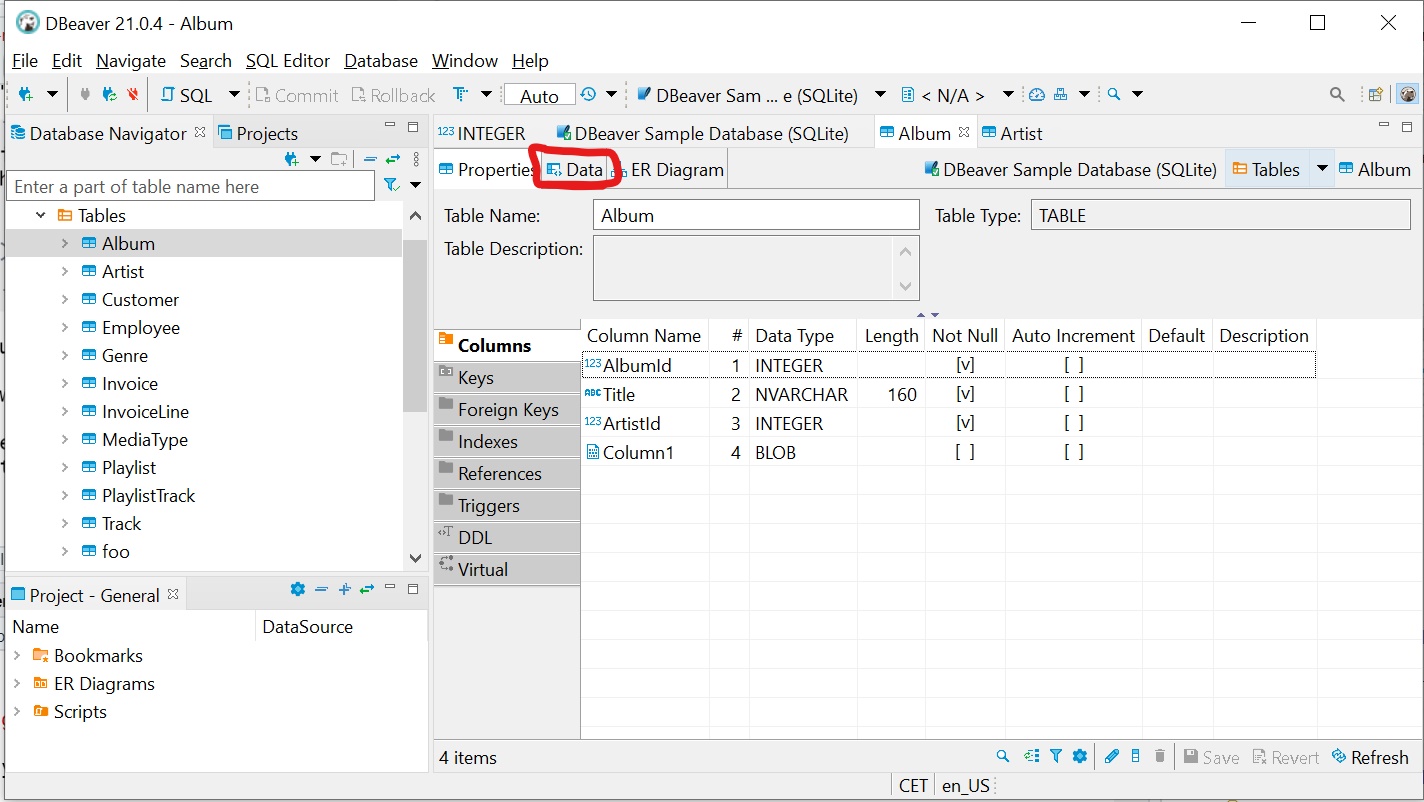
Figure 5.3: The Data tab can be a bit hard to find..
There indeed seems to be a direct link to the Artist table. Double click the Artist table and have a look at that data.
Exercise 5
Which artist has ArtistId 168?
Click for the answer
Youssou N’Dour1
This worked fine, but you had to scroll all the way to number 168 manually. As you know by now, we are notoriously lazy when it comes to having to click or scroll to access data.
When using R, the data tables could for instance have been stored in a package called sample_database. You would load the database you’re looking for, and ask for a specific datapoint: within table Artist on the row where the variable ArtistId is 168, what is the value of variable Name :
(Hypothetical example code, don’t run this, as there is obviously not really a package “sample_database”:)
So in pseudocode (what?)
load database
table %>% filter row %>% select columnIn SQL this looks like (this is actual SQL):
SELECT Name FROM Artist
WHERE ArtistId=168;Or in pseudocode:
SELECT value_from_column FROM database
WHERE filter_on_rows;Some database systems require a semicolon at the end of each SQL statement, so we will do this. SQL doesn’t mind blank lines or spaces. As you can see, the order of the code is different from tidyverse piping.
Select -> table -> filter instead of table -> filter -> select
In DBeaver, click on SQL Editor in the top menu (next to File, Edit, etc) –> New SQL Script. Copy-paste the SQL code above in the SQL script (the actual SQL, not the pseudocode), select it and press ctrl+enter. Does it work?
Exercise 5
Alter the code a bit to see the Albums in the database by the artist with ArtistId 90.
Click for the answer
SELECT Title FROM Album
WHERE ArtistId=90;5.4 Creating a PostgreSQL database in DBeaver
Now that you have played around in DBeaver for a bit, let’s make our own database. If you make a database, it needs to be stored somewhere. One of the more familiar places to store databases is with cloud providers, such as Google Cloud, Amazon and Azure. But actually, just any accessible space is fine. You can store databases on your own computer, or on any server you have access to. Setting up a database on your own computer is free and easy, so this is what we will use in this lesson.
First, download PostgreSQL here.
We will set up our own local PostgreSQL server, and it will only be accessible from the same computer it is installed on. By default, PostgreSQL database server remote access is disabled for security reasons and we will definitely keep it that way.
Run the installer with all the default settings. Remember your password.
(May we recommend using a password manager here if you are not already using one?)
Go back to DBeaver. Click on the icon for “new database connection” just below “file”
Figure 5.4: New connection right there
Figure 5.5: Choose PostgreSQL
Download drivers if DBeaver wants you to.
Now leave all the defaults, but fill in your password:Figure 5.6: Leave all the defaults here.
Figure 3.1: Click on Show all databases
A new connection will have appeared below your DBeaver Sample Database connection, called postgres. This is the default administrative connection database. (If it is all not working, are you sure you actually installed PostgreSQL here?)
Let’s make a new database.
Open a SQL console (DBeaver, like Rstudio differentiates between scripts (code you want to save) and console (other code)). Make sure your new connection is selected. Right click it -> SQL Editor -> Open SQL Console. The new console tab should say "<postgres> Console" on top. Type and run:
CREATE DATABASE myfirstdb;Figure 5.7: Just after creating a new database
In the lower right quadrant of the screen, DBeaver tells you that it made a database. However, it doesn’t yet show up in the left, below postgres.
We can have it show up by refreshing the list on the left: select the connection in the list on the left (postgres - localhost:5432) and press F5 (or right click –> refresh).
But we can also ask using SQL in the SQL console, try this:
SELECT datname FROM pg_database
WHERE datistemplate = false;(In case any of you already knows some SQL: SHOW DATABASES works in MySQL, but not in PostgreSQL.)
We made a database! An empty one, but still a database.
Check if it is indeed empty by running the following code in the SQL console to list all the tables in the database:
SELECT table_name
FROM information_schema.tables
WHERE table_schema = 'public';Exercise 5
Open a new SQL console for the DBeaver Sample Database (right click on the connection name in the list on the left -> SQL Editor -> open SQL console). Copy paste the code to list the available tables and run it. Why does this give an SQL error?
Click for the answer
Because this database is SQLite, not PostgreSQL. There are slight differences in the SQL dialects.
Just as an addition, you did not need to guess this, but in SQLite this translates to:
SELECT
name
FROM
sqlite_master
WHERE
type ='table' ;Check if you see the similarity with the PostgreSQL syntax.
5.5 Filling databases
To keep things clear, we will use ALL CAPS for all SQL functions, as is quite commonly done with SQL.
DBeaver may change text to lower case when typing, that’s no problem. We’ll just use upper case in the reader for clarity.
5.5.1 Creating tables
Now, we will put some data in this database. Again, open a new SQL script or console, but make sure to do so by right clicking your new database in the list now, as the console will take this as it’s working directory.
We will make two, related tables:
CREATE TABLE konijn (
verfnummer int, -- you can put comments here. I have no further comments
oorkleur char(50),
primary key (verfnummer)
);
CREATE TABLE rat ( -- painting rats, no regrets
verf int,
staartkleur char(50),
foreign key (verf) references konijn (verfnummer)
);
Note that we restricted values for “oorkleur” and “staartkleur” to 50 characters. We could assign more here, if you feel like you need more space.
Tables in PostgreSQL databases are organised into schemas. Schemas are analogous to directories in windows, except that you cannot put schemas within schemas. This seems like a bit overkill at first, coming from the SQLite example where we found tables directly under “tables”. But consider having to place everything on your home computer in “My Documents” without further subfolders for work, study, your Pokemon collection and anything related to folk dancing. It will be quite hard to find anything.
(Fun fact: my father, who is in his 70’s actually sort-of does this. He will save documents where-ever his computer suggests it first, including /program files, /windows/addins, anywhere. If he needs to find anything, he just types the name of the file in the search bar. I haven’t been able to convince him that this may not be a good idea… I hope this will convince you though.)
So, your new tables can be found in myfirstdb/Schemas/public/Tables. Go ahead and find them in the list on the left.
Figure 5.8: Here are the new tables.
Double click on one of the tables and check in the ER Diagram tab (next to the Data tab) whether they are indeed related.
We want to keep things a bit organised though, so let’s delete them again. try:
DROP TABLE konijn;This would normally work, but not this time, as our table “rat” depends on “konijn”. Let’s drop them both:
DROP TABLE konijn, rat;or drop “konijn” and all dependend tables like this:
DROP TABLE konijn CASCADE;
Be careful with dropping stuff! You can’t just take them out of the recycle bin like in Windows or MacOS.
5.5.2 Creating tables within a schema
Now we will make a new schema to recreate our tables in (select your database and press F5 after to see it turn up in the list):
CREATE SCHEMA verfexperiment;To create a table within a schema, you just put the required schema in front of the table name. So to make a new table within /public:
CREATE TABLE public.test (
a int,
b int
);
But note that the default is also /public. So if you do not define which schema a table needs to go, it will be put in /public.
Exercise 5
Create the “konijn” and “rat” tables within the verfexperiment schema, but call them “rabbit” and “rat”. Create a third table as well, with whatever variables you like.
Exercise 5
Here is the generic PostgeSQL code for changing the schema name:
ALTER SCHEMA schema_name
RENAME TO new_name;Change schema “verfexperiment” to “paintexp”. After doing so, select the connection in the list on the left (postgres-localhost:5432) and press F5 to see the change.
5.5.3 Data types
As you will have noticed in the examples from the previous paragraph, when you create a table in SQL, it wants you to specify the data type. For example:
CREATE TABLE public.test (
a int,
b char(8)
);
We used int (integers) and char(n) (character) before, with the latter even asking you to specify the maximum length. There are a lot of possible possible data types in PostgreSQL (check them here), most notably:
- int: the typical choice for whole integer data
- decimal(precision, scale): define how many digits (precision) and how many of them after (scale) the decimal point. So the number 12.3456 has a precision of 6 and a scale of 4.
- float: also decimals, but limited precision. Faster to work with.
- char(n): characters, fixed length
- text: characters but unlimited length (this takes more space)
- boolean: TRUE/FALSE
Storing numbers in databases can be a bit tricky. Excel for example stores numeric data with a maximum of 15 significant digits. You may have run into this before. For example, see the following image from Wikipedia:
Figure 5.9: Example of precision issues in Excel By Brews ohare - Own work, Public Domain
The top figure shows what Excel will show for 1/9000 in red. While this number will have an infinite string of ones, only the first 15 ones will be stored (leading zeroes don’t count). In the second line, the number one is added to the fraction, and again Excel displays only 15 figures (red). Now if we subtract 1 again from z (third line), mathematically, the result should be the same as the top line. However, it is not. It is not even what you would expect to get from the numbers displayed, as you would have expected to get z-1 = 0.000111111111110000. So the numbers excel is showing you are not the numbers it is doing calculations with. This is not a bug in Excel, it is an issue with how computers are designed in general.
Try the same calculations in R:
We won’t go into detail with floating point problems and binary calculations, but remember that if you ask a computer to remember “1/9000” for you, it won’t store exactly 1/9000.
In SQL, you can define however how precise it needs to be. The data type decimal/numeric (they’re the same in PostgreSQL) can store numbers with a very large number of digits and perform calculations with them. So if you require the numbers to be stored exactly (such as monetary amounts), use this data type. If you don’t need numbers to be rounded to an exact precision and have a lot of data to work with, you can use float instead of decimal, as it is faster.
5.5.4 Generating data with SQL
Now we will put some data into our rabbit and rat tables.
insert into paintexp.rabbit
(verfnummer, oorkleur)
values
(1, 'red'),
(2, 'blue'),
(3, 'green');
insert into paintexp.rat
(verf)
values
(2),
(3);
Have a look what happened in both tables (press F5 if you don’t see anything).
Exercise 5
Add 2 more rows in both tables.
Exercise 5
Verify with SQL what’s in the tables now, here is the generic SQL code:
SELECT * FROM table;Click for the answer
SELECT * FROM paintexp.rabbit;
SELECT * FROM paintexp.rat;5.5.5 Altering tables
We made a column for the rat tail colours, but it is empty. If we want to change this, we need to update the table.
UPDATE paintexp.rat
SET staartkleur = 'orange'
WHERE verf = 3;Exercise 5
Suppose you have a table with 50 rats and want to change a specific rat’s tail colour value? What would you need to be able to use the code from the example above?
Click for the answer
A unique identifier for each rat. So basically, a simple key.The PostgreSQL ALTER TABLE command is used to add, delete or modify columns in an existing table.
ALTER TABLE tablename ADD column_name datatype;Exercise 5
Add a column to the rabbit table named ‘rating’. You can leave it empty
You can also use ALTER TABLE to change the table name, in which case the schema needs to be specified in the old name, but not the new:
ALTER TABLE paintexp.rat RENAME TO ratsOr if we for instance want to add anything to the text in a column. Try what the following query does:
update paintexp.rabbit
set oorkleur = 'brownish '||oorkleur;Before we continue, why are we doing this again? This seems hard? What’s wrong with typing some data into Excel? Or even Notepad? Well, that depends. If you have just a little data, typing it manually and storing as a .csv is fine. But most large databases out in the wild are accessible through SQL. Also, suppose you want to add some data to such a database. Using SQL to do so, ensures that you are working reproducible, as you are using code to change anything.
5.6 Filtering and selecting
SQL is often used to find specific data in a database. We will practice a bit with another example database: pagila. Pagila is a “DVD Rental Store” example database for PostgreSQL which is not very biological, but it is specifically designed to show case features within PostgreSQL.
Download Pagila here. Pagila is an example database.
Make a new database called pagila.
Figure 5.10: The new database on the left
Open psql (SQL shell, Psql is the interactive terminal for working with Postgres) and enter the following (add your password). (If you have no clue where psql.exe is, just type “psql” in the search bar in Windows.)
Figure 5.11: Screenshot of psql
It should say pagile-#.
Type (change “yourpathhere” to reflect where you put the files):
\i yourpathhere/pagila-schema.sqlFor me, this looks like this
Now type (again, update the path):
yourpathhere/pagila-insert-data.sql
And wait for it to finish.
Go back to DBeaver and refresh the connection, and our pagila database will be filled! Also, you have met psql, PostgreSQL’s shell thingy, in the process.
If for some reason some tables are empty, try in psql:
yourpathhere/pagila-data.sql
Figure 5.12: We have data!
5.6.1 Selecting
This database contains fictional business processes of a DVD rental store. It contains 15 tables. We will use it to try some queries (a query is a request for data results from a database). In fact, we already did some queries, such as:
select * from database.table; Is in fact sending a query for all columns in a table.
You can also ask for specific columns.
If this doesn’t work, type the following, press TAB after actor to get suggestions on column names. Some letter will appear.
SELECT
first_name,
last_name
FROM
actor;Exercise 5
Select only the customer_id column from the customer table.
The following example uses the SELECT statement to return full names and emails of all actors, using || as the concatenating operator that concatenates one or more strings into a single string, and using AS to give the new column a descriptive name. Try what happens if you omit the “AS full_name”.
If this doesn’t work, type the following, press TAB after actor to get suggestions on column names. Some letter will appear.
SELECT
first_name || ' ' || last_name AS full_name
FROM
actor;You can order your selection, in ascending (ASC) or descending (DESC) order.
SELECT
title,
release_year
FROM
film
ORDER BY
release_year DESC,
title ASC;Exercise 5
What did the previous query do?
Click for the answer
Select the columns title and release_year from the table film and sorts the rows first by the release year in descending order and next by the title in ascending order.Which actually did not make much sense, as all releases were in 2006:
SQL has a unique() equivalent as well: DISTINCT. Use it to check if indeed all release_years were 2006:
SELECT
DISTINCT release_year
FROM
film;Now let’s see which actor has the longest first name:
SELECT
first_name,
LENGTH(first_name) len
FROM
actor
ORDER BY
len DESC;5.6.2 Filtering
If you do not want to get the whole column, but a specific set of rows, you can filter. In SQL, this is called WHERE:
SELECT select_list
FROM table_name
WHERE conditionTidyverse filter(columnA==4) would translate to WHERE columnA=4
Exercise 5
Find all the actors named ED, and have them returned to you in alphabetical order of their last name.
Click for the answer
SELECT first_name, last_name FROM actor WHERE first_name='ED' ORDER BY last_name ASC;
You can search for patterns within a column with the LIKE operator. Read about it here
Exercise 5
Find the actors with a first name starting with the letter B, sort them in alphabetical order and return their full name.
Click for the answer
SELECT
first_name || ' ' || last_name AS full_name
FROM
actor
where
first_name LIKE 'B%'
ORDER BY
first_name ASC;We can add operators like AND, OR and IN as well, just like in tidyverse filtering, as well as !=
SELECT
first_name,
last_name
FROM
actor
WHERE
last_name = 'HARRIS' OR
first_name IN ('BOB','BEN','Bill');
Now that we have introduced you to the W3schools website, you can actually look up the syntax for all kinds of operations in the menu on the left. tutorialspoint also has a good list. Use these two websites to solve the following questions:
- Which customers have a first name BETWEEN 5 and 7 characters long?
- What was the MAX amount payed for a rental?
- Find all actors not called ED.
- COUNT the number of films in the database.
- Find the last names of all actors and customers (UNION)
- Play with the SQL quiz if you like.
5.6.3 group by
The GROUP BY statement groups rows that have the same values into summary rows, like “find the number of films in each category”. Try this:
SELECT COUNT(film_id), category_id
FROM film_category
GROUP BY category_id;And run this code to list the last names of actors and the number of actors who have that last name, but only for names that are shared by at least two actors.
SELECT
last_name
,COUNT(last_name) AS last_name_count
FROM actor
GROUP BY last_name
HAVING COUNT(last_name) >= 2
ORDER BY last_name_count DESC
;
5.6.4 Joining
But now what if we want to combine information from different tables?
On the bottom of this page is an image explaning the different kind of joins with their SQL syntax. Note that the image does not show you what happens when there is missing data.
You could for instance want to display the first and last names, as well as the address, of each staff member, but this information is in two separate tables:
SELECT
first_name
, last_name
, address.address AS street_address
FROM staff
INNER JOIN address ON address.address_id = staff.address_id
;Exercise 5
What does “INNER JOIN address ON address.address_id = staff.address_id” mean? How are these attributes called? What is the underlying assumption with using these keys?
Click for the answer
This part does an inner join, selecting only records that have matching values in both tables, with address_id in staff as a primary key and within address as a foreign key. It assumes that no two (or more) staff members live at the same address.Or add all the category information to each film title in the film table:
SELECT
*
FROM
film
LEFT JOIN film_category
ON film_category.film_id = film.film_id
LEFT JOIN category
ON category.category_id = film_category.category_id
ORDER BY title;5.7 from PostgreSQL to somewhere else
Exporting your query results to .csv is quite easy:
- right click anywhere in the SQL code of the query you want to export and choose Execute > Export from query. (alternatively, right click on the result and choose Export Data)
- Choose CSV
- Leave all the default settings, but choose a destination folder and if you like, change the file name.
Exercise 5
Make a table containing only all the film titles and their category (Documentary, Horror etc..). Export this as .csv
5.8 From R to PostgreSQL
The RPostgres package contains a database interface and ‘PostgreSQL’ driver for R.
Start with installing some packages:
install.packages('RPostgreSQL')
#install.packages('devtools') # you should have this one already
install.packages('remotes')
install.packages('RPostgres')And make a connection to the PostgreSQP server again, but now from R (fill in your password):
library(DBI)
con <- dbConnect(RPostgres::Postgres(),
dbname = "pagila",
host="localhost",
port="5432",
user="postgres",
password="password") Use the following code, what does it do?
Also try:
And
You can actually write SQL queries as well:
dbGetQuery(con, 'SELECT * FROM actor')In RMarkdown, you don’t even have to do the dbGetQuery() function, you can just insert a SQL code chunk (however, both starting the connection and closing it again still needs to be done in r code chunks.)
'''{sql, connection=con}
select * from actor where actor_id > 75;
'''Or if you prefer: make a connection to a table and use tidyverse syntax to send queries.
Try if this works and send a few queries! One example to get you started:
5.8.1 Parameterized query
It is very tempting to create queries with variables by pasting strings together:
# DON'T DO THIS!
myvar <- "bla"
dbGetQuery(con, paste0("SELECT * FROM table_name WHERE column_name = '", myvar ,"'"))But this will pose a security risk if you do this on connections over the internet. Someone may alter the query to contain more rows than you intended or even damage the database:
# EVIL ATACKER BE LIKE:
myvar <- "bla'; DROP TABLE 'table_name"
dbGetQuery(con, paste0("SELECT * FROM table_name WHERE column_name = '", myvar ,"'"))Instead, use a parameterised query with dbSendQuery() and dbBind():
You create a query containing a placeholder and send it to the database with dbSendQuery(), Use dbBind() to execute the query with specific values, then dbFetch() to get the results. $1 is the placeholder here, and we substitute it with a 4:
myquery <- dbSendQuery(con, "select * from actor where actor_id = $1")
dbBind(myquery, list(4))
dbFetch(myquery)When you’re done with the results from a prepared query, clear the result:
Let’s disconnect and connect to the myfirstdb database:
dbDisconnect(con)
con <- dbConnect(RPostgres::Postgres(),
dbname = "myfirstdb",
host="localhost",
port="5432",
user="postgres",
password="password") And write some data to the database from R:
To write for instance the penguin data to our myfirstdb database:
Go to Dbeaver and check myfirstdb -> schemas -> public -> tables to check if it worked.
Exercise 5
Actually, we had better uploaded tidy data.. Remove the table again with
Transform the penguins data to tidy data format (you actually already did this in the first part of this lesson) and upload it to the myfirstdb database.
Click for the answer
And finally, the connection must be closed:
Portfolio assignment
Maak opdracht 5 van de portfolio-opdrachten.
5.9 Resources
https://db.rstudio.com/best-practices/run-queries-safely/ https://nuitrcs.github.io/databases_workshop/r/r_databases.html
5.10 I really love SQL and want more
There are a lot of exercises here at pgexercises.com/
And here as well
Here are some [more queries for the pagila] database(https://github.com/Jkremr/pagila-queries/blob/master/README.md)
Here is a blog post on building complex queries
CC BY-NC-SA 4.0 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. Unless it was borrowed (there will be a link), in which case, please use their license.
I absolutely chose this example artist to tease the other teachers, as they will probably have a certain song in their heads for the rest of the day.↩︎