Lesson 3 Data management
Picture by Hamid Hajihusseini, CC BY 3.0, via Wikimedia Commons
3.1 Aim of this lesson
After this lesson you will be familiar with agile working:
And with the most important data management guidelines and principles:
- Proper file naming strategies
- Guerrilla Analytics principles
- Files and folders / Project structure
- Checking data validity/integrity using checksums
- Data-formats & Data shapes / Tidy data
- metadata
- Encoding variables & Exploratory Data Analysis
- Preparing your data for sharing
- Proprietary vs non-proprietary formats
3.2 File names
Do you recognize this?
 https://medium.com/@jameshoareid/final-pdf-finalfinal-pdf-actualfinal-pdf-cae61ab1d94c
https://medium.com/@jameshoareid/final-pdf-finalfinal-pdf-actualfinal-pdf-cae61ab1d94c
And this?

The use of version control abolishes the need for inventing a file name every time you save it. You will learn more about using version control (git and github.com) in lesson 2 and 3. With the use of git version control you only have to think about naming a file just once with a good name. But what entitles a ‘good’ file name?
A good file name is:
- Unique in a folder (prevent duplicated names)
- Is short, but descriptive (if you need it to be longer to be descriptive enough, choose that)
- Does not contain any special characters* except for
_and a.before the extension. Having multiple dots (.) in a file name can be confusing but sometimes is required. For example for an archive we sometimes see<file_name>.tar.gz - The typeface of a file name is ideally set in lowercase only. If you want to deviate from this use
UpperFirstcamelcase instead. - The most important thing about naming files is to be consistent. This is also the hardest part!
- If you receive a file from somebody else: Never change the file name, even if it does not meet the above requirements. Changing a file name causes a breakage between the file and the source it came from. If you change a file name you recieved from a person or downloaded from the internet, the person who send the file will not know about the new name.
*The special characters you should avoid in a file name:
! @ # $ % ^ & * ( ) + - = : " ' ; ? > < ~ / ? { } [ ] \ | ` , Special characters are reserved for other purposes and can cause problems when a back-up of the files is made or when files need to be loaded in analyzing software or when copying files.
**Basically, what was stated about file names, also applied to naming variables in a dataset. Or, with other words: choosing or creating valid names for columns in a data frame, or names of R objects for that matter.

Below, I show an example of badly formatted file name and column names to make the point.

Exercise 3
what is wrong with this file name and its headers? can you spot another problem with the data sheet?
3.3 The Guerrilla Analytics Principles
To help you build a thorough data management process for yourself that you can start using and expanding when needed, we need a framework. In this course we use the Guerrilla Analytics framework, as described by Enda Ridge in this booklet. If you are pursuing a career in Data Science, I highly recommend getting a copy! This booklet describes in a very practical, and hands-on way, how to establish a data management workflow. Either when you work all by yourself or in a team, the pointers in this book will be applicable in both situations. Once you get to know this framework, you will discover that you used to do it wrong (like me…).
To build the framework, let’s look at the core 7 Guerrilla Analytics Principles:
- Space is cheap, confusion is expensive
- Use simple, visual project structures and conventions
- Automate (everything - my addition) with program code
- Link stored data to data in the analytics environment to data in work products (literate programming with RMarkdown - my addition)
- Version control changes to data and analytics code (Git/Github.com)
- Consolidate team knowledge (agree on guidelines and stick to it as a team)
- Use code that runs from start to finish (literate programming with RMarkdown - my addition)
Guerrilla Analytics book by Enda Ridge,

As you can see from my own edits to these principles, quite a few are immediately applicable when using a programming Language like R and its Rmd format, that we use all the time. I will go over each principle below in more detail. But first an exercise.
Exercise 3
Imagine you receive a file attached to an email from a researcher in your research group called:
salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx
The file is located in the course ./data folder.
You are requested to run an analysis on the raw data in this file (sheet named ‘All Cycles’). It contains data from a plate reader experiment where wells are measured over time. The researcher asks you to generate a plot per sample. No other information was provided in the original email.
Describe the following steps in an RMarkdown file. You do not need to write the R code for the actual analysis at this point - we will do that later in another exercise in this lesson. Answer these questions in your markdown file with exercises.
- How would you prepare for this analysis in R/Rstudio?
- Look at the contents of the file, which Excel Worksheet do you need for the analysis?
- Which steps do you need to take to load the data into R
- Which steps do you need to take in reshaping the file to make the dataformat match the requirement for using `{ggplot}
- Think of a better file name
- Write a conceptual and kind and friendly, but clear reply email (in English) to the researcher, where you address the changes that the researcher needs to make to the file in order for you to be able to analyze this data in R.
Now that we encountered a data management challenge in the Wild, let’s build our framework to be able to tackle these types of problems in a more structured fashion, next time we meet them.
3.4 The Guerrilla Principles explained
3.4.1 Principle 1: Space is cheap, confusion is expensive
This principle is simple: storage costs are low these days so there is no need to spend a lot of time on administrating files.
- Keep your files, you never know when you need them. The price for a storage units has dramatically dropped over the past years, so there is no need to delete files or clean up any more. Just save old files in an archive.
- Store data that you actively work on in online-cloud storage. Usually, you will be better of storing data on a remote location in the cloud. This reduces the risk of data loss, when something happens to the local file system. Cloud storage is usually redundant. Which means copies of the same file are ditributed over mupltiple locations (even geographically if you want). When one disk fails, there is always a back up, so no real data loss if hardware dies.
- Protect yourself: do not click on attachments and spiffy emails, cyber criminals are getting smarter everyday.
- Create md5sums for important (source) data-files to ensure data integrity and data transfer validity. A MD5 ‘hash’ or ‘checksum’ is a 128-bit summary of the file contents. Files with different MD5 sums are different. So you can use this to check whether a file has changed since you last used it.
- Agree on a storage system, share it, use it, stick to it

Data integrity
When receiving a file from a laboratory that has performed a sequencing analysis, the files you receive are usually in .fastq.gz or fasta.gz format. Because these files can be big, they are usually accompanied by a small file containing a hash-like string looking something like this:
e785f3f3dc9bdcb6c6048e7f883c1bfb
This is the md5sums checksums for the file used earlier: ./data/salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx. When the file changes the checksums also changes, like we will see in the following exercise.
There are a number of different algorithms with which we can calculate such so-called sumchecks. Here we use the md5sums, which is a popular hashing algorithm.
md5sums are
- A unique code to identify a file
- Can be used to verify the integrity or the version of a file
- Can be genarated from Windows, MacOS, Linux or from within e.g. R/Python/Bash
- md5sums are also used for safety: checking an md5sum ensures that the code is valid and has not changed (e.g. Anaconda)
- There are many different types of hash functions MD5, SHA256 are much used for data and software
In the following example, we find the MD5sum for a .txt file in /data/md5_examples:
library(tools)
md5_ex1_raw <- tools::md5sum(
here::here(
"data",
"md5_examples",
"MD5_exampledata_1.txt"
)
)Use enframe() to get atomic vectors or lists nice and tidy in a tibble:
Exercise 3
There are actually 4 very similar files lying around.
Find out which one is different from the other three using their MD5sums.
If you want, md5sum can handle multiple files at the same time, but wants full paths for all of them.
Click for the answer
#library(tools)
myDir <- here::here(
"data",
"md5_examples2")
fileNames <- list.files(myDir, recursive = TRUE)
tools::md5sum(file.path(myDir, fileNames)) %>% enframe() -> md5sums_all
md5sums_all$filename <- fileNames
md5sums_all %>% select(filename,value)## # A tibble: 4 × 2
## filename value
## <chr> <chr>
## 1 MD5_exampledata_1.txt f3d2eeb79b4c24490bf08f483e5cadd3
## 2 MD5_exampledata_backupcomputercrash.txt f3d2eeb79b4c24490bf08f483e5cadd3
## 3 MD5_exampledata_maybethisone.txt a331f23fd3586f0100173bd28b1e319c
## 4 MD5_exampledata_savethisone.txt f3d2eeb79b4c24490bf08f483e5cadd3Exercise 3
Calculating and checking md5sums checksums in R
For the data in this exercise use the file toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.csv
toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.csv
- Determine the md5sums of the file. Save the checksums to a file [write a piece of R code]
Click for the answer
## (a)
# calculate md5sums for file "toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.csv"
tools::md5sum(
here::here(
"data",
"toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.csv"
)
) %>%
enframe() -> md5sums_toxref
md5sums_toxref %>% # write as dataframe for easy access
readr::write_csv(
here::here(
"data",
"toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.md5")
)- Upload the file to the Rstudio server and check again. Again, save the resulting md5sums in a file. Do the md5sums you calculated on the server and the md5sums of the local file match?
Click for the answer
You should get 399276ed77401f350aaee0c52400c5e9 as md5sums on both the server and locally- Download the md5 file you just generated on the server (or upload the one you generated locally). Can you think of a way to check in an R script whether they are the same? Write the script and save it in a .R file
Click for the answer
# calculate checksums from local file as under (b) and write to file
# calculate checksums from local file as under (b) and write to file
# From RSTudio
# load both md5sum files into R
# you could just logically compare the two srings, but is there a better way?
serverside <- read_csv(
here::here(
"data",
"toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE.md5"
)
)
local <- read_lines(
here::here(
"data",
"toxrefdb_nel_lel_noael_loael_summary_AUG2014_FOR_PUBLIC_RELEASE_local.md5"
)
) %>%
enframe()
## check equality
serverside$value == local$value[2]## [1] FALSE3.4.2 Principle 2: Simple, visual project structures and conventions
This principle concerns the organisation of files and folders on the file system. Because you usually do not work alone, but in a team, this is an important point to agree upon within your team. If you do not stick to the rules you agreed on playing by, you will find that playing the game is no fun at all. When organizing a classical project structure, people with no data science background tend to organize files and folders on the contextual, or categorical level. This means you would have something like this:

This makes sense but has at least two major drawbacks:
- Deep nesting of files in folder causes the absolute path to a file to grow. The path length on some file systems (especially in the cloud like OneDrive or GoogleDrive have a limit to the length of a path size. When copying a file or folder, this may cause the file or folder not be copied. Usually this error is silent. Meaning, you will not know about it, before it is too late and you discover years later that a complete folder is empty - trust me, it happens, I’ve been there!)
- Categories and projects evolve. So at the time of putting the structure in place this may seem like a logical structure, but things will change. There will be new categories and new types of files, you had not thought of before. Where will these go?
See for my own bad example on how not to do it:
## /Users/bas/Desktop/git_repositories_readers/tlsc-dsfb26v-20_workflows/wrong_structure
## ├── Data files 001
## │ ├── Final Results
## │ │ └── experiment_1_results_final.txt
## │ └── experiment_1.txt
## ├── Manuscripts
## │ ├── me_et al , 2020_v01 - Copy.docx
## │ ├── me_et al , 2020_v02.docx
## │ └── me_et al , 2020_v03_final_final.docx
## ├── Project Documentation
## │ └── applications
## │ ├── Application final prject x.docx
## │ └── application_final_project y.docx
## └── Volunteer responses
## ├── Patient 2.xlsx
## ├── Patient_3.xlsx
## └── patient_1.xlsxSo when managing files in projects adhere to the following guidelines
- Create a separate folder for each analytics project . Keep the unit of a project small. So a larger research project can usually be subdivided into smaller projects. Create an analysis project for each sub-project. Don’t try to fit it all together.
- Do not deeply nest folders (max 2-3 levels, so no project/documentation/applications/metc/first_communication/input/stuff/morestuff/mail.txt)
- Keep information about the data, close to the data. So store descriptions about variables for example in a README.txt file and store that README.txt file in the same folder as where the data lives that it describes. When using Excel files, you may choose to store the README information in a separate worksheet. More on this later 3.5
- Store each dataset in its own sub-folder. I usually work like the example below. Each dataset I receive goes in a numbered folder in the
data-rawfolder. If I recieve an updated file, I add the old file to a new folder calledv01inside the orginalData..folder. The updated file replaces thev01file. If then you recieve yet another update, you can repeat this trick with av02folder inside theD..folder. In this way, the file directly in theD..folder is always the latest updated version. The README.txt file containing data about the data (see 3.5) lives also in the root of theData..folder. - Do not change file names or move them. As explained before, never change a file name of a file you recieve or download from the internet. If you need to change the file for analysis reasons (sometimes bioinformatics pipelines require specially formatted file names and than you do not have a choice, but to rename them), record your changes in the README.txt file!
- Do not manually edit data source files. in fact Never, ever change a value inside a file. You can (almost) always solve this using code.
- In code, use relative paths (here::here etc)
- Use one R project per project (in RStudio -> RStudio Project) Don’t work outside of projects, and don’t reuse projects for other stuff.
How to organise data files
## /Users/bas/Desktop/git_repositories_readers/tlsc-dsfb26v-20_workflows/data-raw
## ├── Data010
## │ ├── 2020-06-19_covid_ecdc_cases_geography.csv
## │ ├── 2020-06-19_md5sums_covid_ecdc_cases_geography.md5
## │ ├── README.txt
## │ ├── supporting
## │ │ ├── covid_ecdc_cases_geography.R
## │ │ └── md5sums.R
## │ └── v01
## │ ├── 2020-05-31_covid_ecdc_cases_geography.csv
## │ └── 2020-05-31_md5sums_covid_ecdc_cases_geography.md5
## └── Data020
## ├── README.txt
## └── messy_excel.xlsx3.4.4 Principle 3: Automate everything with program code
If we consider this lesson is about Reproducible Research, this is the core principle we need to consider thoroughly. From the R programming perspective, we can do this easily when using RMarkdown in combination with scripts. It already automates everything from the data file to the final results of an analysis. And, what is even better: it is neatly packaged in a reproducible report that, when rendered to html, can be opened on every computer.
Take heed of these pointers:
- Do everything programatically (in code) for reasons of reproducibility
- Store clean curated datasets in the “data” folder, with md5sums and a README.txt
- Use literate programming (RMarkdown or Jupyter Notebook) for full analysis
- Store scripts in a “./code” or “./inst” folder
- Store (R) functions in R in a “./R” folder
Exercise 3
Let’s demonstrate this principle 3 with a COVID-19 reporting example
- Imagine we want daily reports on the number of COVID-19 cases and caused deaths
- We want to be able to dynamically report data for different countries and dates to compare situations in the World
- The data is available (for manual and automated download) from the European Center for Disease Control
- The analysis can be coded completely from begin to end to result in the information we need
Take a look at the source file in this Github repo (click)
Download this file to your RStudio environment and knit the file.
What do you see in the rendered html?
What happens if you change some of the parameters in the yaml header of the file, in particular country_code or from_to_date?
Parameterization
The above example in the exercise is an example of a so-called parameterized script. In this case a parameterized RMarkdown. We will learn more on parameterizing RMarkdown files in lesson 9
- The covid Rmd is parameterized on date and country
- The script automatically includes the parameters in the title of the report and the captions of the figures
- The ‘rendered’ date is automatically set, for tracking and versioning purposes
- Parameterization can used to automate reporting for many values of parameters
- Further automation is easy now (although the ECDC has regular ‘changes’ to their latest data available for download - and they do not use md5sums!! - This makes full automation and building–in checks more difficult)

3.4.5 Principle 4: Link stored data, to data in the analytics environment, to data in work products
Read this paragraph header again and make sure you understand the difference between “stored data”, “data in the analytics environment” and “data in work products”.
Basically this is what you are doing with literate programming (e.g. RMarkdown) with R or Python in RStudio or Jupyter:
- The data is stored on disk or in the Cloud
- The
analytics environmentis the Global Environment (where variables and R-objects live) - Data is pulled from the storage in the Analytics Environment by a script
- The work products (Rmd / Notebooks) bring it together
3.4.6 Principle 5: Version control for data and code - Git/Github
- When you do data analysis, you should use code. See also Principle 4.
- When you write code, you should use Git, preferably in combination with Github. Or use another version control system.
- Hence: When you do data analysis, you should use Git & Github
- Git/Github is ‘track-changes for code’
You will learn more on using the git/github workflow in data science in later lessons (1, 1.7, 2, 2.3).
3.4.7 Principle 6: Consolidate team knowledge
AKA communicate! When working together it is vital to come to an agreement on how you work together. I hope the Guerrilla Analytics framework provides a starting point. Hopefully, you will learn during your projecticum work how vital this actually is when working together on a data project or any project for that matter. Here are some pointers:
- Make guidelines on data management, storage places and workflows
- Agree within the team on them
- Stick to them! And be frank about it when things go wrong or people misbehave. An honest and open collaborative environment is encouraging. It is usually hard for people to change their way of working.
- Work together in a virtual collaboration environment.
- Work together on code using Github or some other version control based repository system (e.g. Gitlab / Bitbucket).
- Provide for education and share best practices within the organization, the department and/or the team (this is what we try to achieve with this course).
Exercise 3
tips
Please share all tips for fellow students on the prikbord in teams!
3.4.8 Principle 7: Prefer analytics code that runs from start to finish
- Create work products in RMarkdown (or Jupyter notebooks if you like)
- Write functions that isolate code and can be recycled. When writing a function, think about how to generalize this function so that you can recycle it in other projects. This saves time and adds robustness.
- Use iterations to prevent repetition. Write clear and compact loops, for example in R by using the map-family of functions from the
{purrr}package. I prefer these above writingfor()loops because they focus on the object that is being changed, not on the computation that is done. - For the future: In R, create an R-package. Once you have a fully functional RMarkdown file, it is quite easy to rebase that code into an R Package. I call this the “Start with Rmd” - workflow. The demo in the link shows you an example, you don’t need to do the demo now, but use it in the future if you like.
3.5 Metadata
Meta data is data about the data, such as for instance the type of variables, number of observations, experimental design and who gathered the data. This is quite often not reliably documented (or at least not easily accessible) but very important: data without context loses some of its purpose.
Take a look at this Wikipedia image of cocoa pods and scroll down. As you can see, Wikipedia stores a lot of metadata on file usage, licence, author, date, source, file size… Even the original meta data from the camera is included (scroll to the bottom).
Meta information for data files, like type of variables, ranges, units, number of observations or subjects in a study, type of analysis or experimental design often goes in a README.txt file or a sheet in the Excel file containing the data. Keep the readme information close to the data file. Also, information about who is the owner of the data or who performed the experiment when and where and with what type of device or reagents is very useful to include. In our exercise above such README information would have saved us a lot of time figuring out what is what in the Excel file, don’t you think?
An example of a readme file is depicted below.
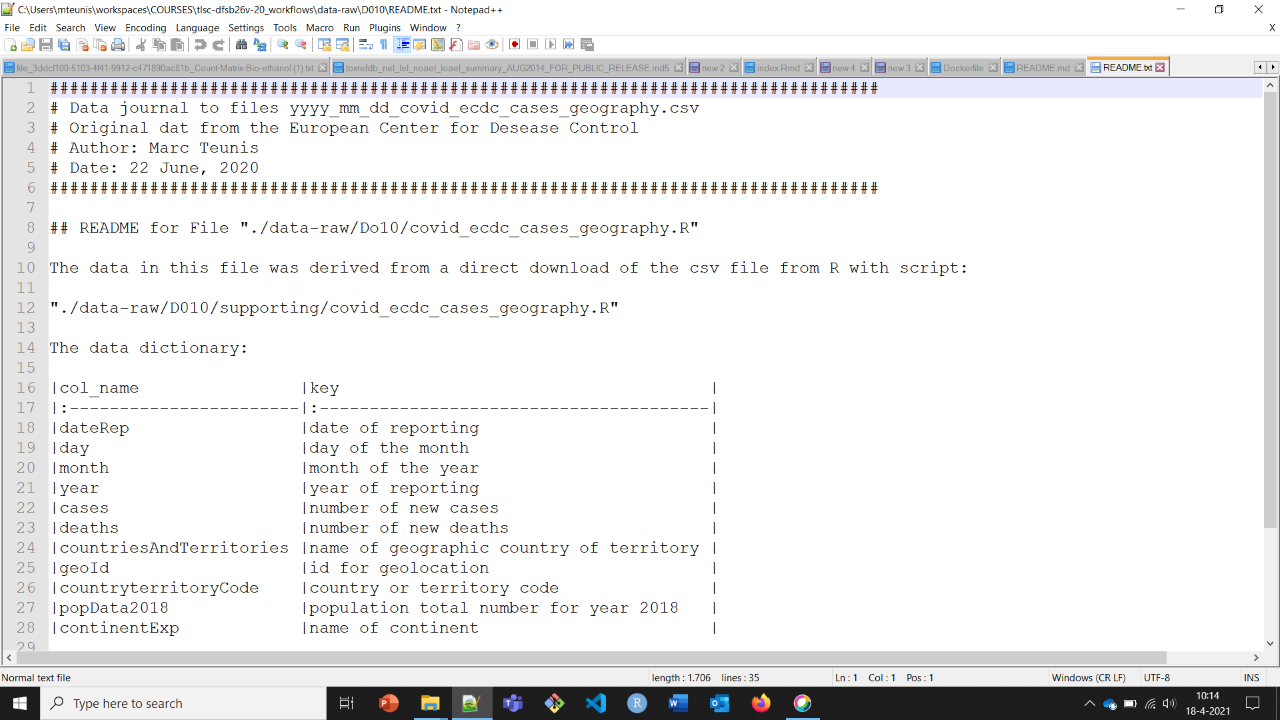
Figure 3.1: A short example of a README.txt file
It does not need to be very long, but provides information on where the data (which project?) refers to, who the owners are, who to contact in case of questions and what are the contents of the data (variable description)
3.5.1 YAML header
Rmd files include a metadata section themselves: the YAML header. At the very least, specify the title, author and date here.
3.5.2 saving metadata
Save at the very least in your Readme.txt:
- General information
- title
- information about the authors (name, institution, email address)
- date of data collection
- location of data collection
- license information
- a data log with for each data file:
- short description
- date the file was created
- variable list (full names and explanations)
- units of measurements
- definition for missing data (NA)
- methodological information
- description of methods (you can link to piublications or protocols)
- description of data processing (link to RMarkdown file)
- any software or specific instruments used
- Describe details that may influence reuse or replication efforts
Here is a nice example file by the university of Bath for bioinformatics projects, another more general template available for download, and here another template for experimental data
This may seem to be a bit too much for your current projects, but try and see how much you could fill in and keep the template for future projects! Remember that any metadata is better than none.
3.5.2.1 Data-log
This is an overview of all datafiles in a project.
Keep an MS Excel file (called “data_log.xlsx”) in the root of the folder \data of each project and keep it up to date to track all the files present in this folder. Provide names and additional information here. Meta information for the \data_raw folder is best kept in that folder in a README.txt file.
Use versioning of data files. Decide on a versioning system for yourself (we showed you an example before, but you are free to look for other options), an stick to it.
3.5.3 Annotations and meta data in multiple files
Some metadata is most useful if you have it available in the code directly.
In order to reduce effort in generating a complete tidy table for your data it might be worthwhile to create a number of extra tables containing meta data. Typically this is how it would work:
Assume we have wide data format originally created in Excel looking like this: (Actually, we will ask R to generate some data that looks like it was imported from excel instead, because we didn’t feel like copy-pasting 4x100 numbers to make an excelfile…)
# generate some dummy data for the example
measured1 <- rbinom(100, size = 2, prob = 0.3)
measured2 <- rnorm(100, mean = 5.3, sd = 0.1)
measured3 <- rnbinom(100, size = 10, prob = 0.1)
concentration <- rep(1:10, 10)
# put it in a tibble
data <- tibble::tibble(
`concentration (mMol/l)` = concentration,
`measured 1 (pg/ml)` = measured1,
`measured 2 (ng/ml)` = measured2,
`measured 3 (ng/ml)` = measured3
)
data## # A tibble: 100 × 4
## `concentration (mMol/l)` `measured 1 (pg/ml)` `measured 2 (ng/ml)`
## <int> <int> <dbl>
## 1 1 0 5.39
## 2 2 1 5.33
## 3 3 0 5.19
## 4 4 2 5.27
## 5 5 2 5.24
## 6 6 0 5.41
## 7 7 0 5.33
## 8 8 1 5.29
## 9 9 0 5.36
## 10 10 0 5.30
## # ℹ 90 more rows
## # ℹ 1 more variable: `measured 3 (ng/ml)` <int>The concentration (mMol/l), measured 1 (pg/ml), measured 2 (ng/ml), measured 3 (ng/ml) are the variable names as provided in Excel. As you can see they are not adherent in a few ways tot the aforementioned naming conventions ( 3.2). The measured 1 (pg/ml), measured 2 (ng/ml), measured 3 (ng/ml) refer to three variables that were determined in some experiment. The units of measurements are (as is common in Excel files) mentioned between brackets in the column name.
For compatibility and inter-operability reasons this data format can be improved to a more machine readable format:
In this case, the unit information that is included in the variable names can be considered metadata. So you can put that information in a separate table. In the example below, I will call it coldata (short for column data)
First we need to create a pivoted table where the first column represents the variable names of our data table. Then we need to add a row for each variable in our data. It is best if the variable names and the values in the meatdata table in column 1 excactly match (in term of spelling and typesetting). I will show how this looks for our data
## # A tibble: 4 × 1
## varnames
## <chr>
## 1 concentration (mMol/l)
## 2 measured 1 (pg/ml)
## 3 measured 2 (ng/ml)
## 4 measured 3 (ng/ml)We now have a metadata table with one column called varnames. However, we are not done. If we want to create a tidy format of our metadata table we need to separate the unit information from the variable names column. Let’s extract the units into it’s own column
metadata %>%
mutate(
varnames = str_replace_all(
varnames,
pattern = " ",
replacement = "")) %>%
separate(
varnames,
into = c("varnames", "units"), sep = "\\(", remove = FALSE) %>%
mutate(
units = str_replace_all(
units,
pattern = "\\)",
replacement = "")) -> metadata_clean
metadata_clean## # A tibble: 4 × 2
## varnames units
## <chr> <chr>
## 1 concentration mMol/l
## 2 measured1 pg/ml
## 3 measured2 ng/ml
## 4 measured3 ng/mlWe can now start adding additonal information such as remarks or methods to the the metadata column.
3.6 Study documentation
The folder \doc contains documentation and can be basically everything concerning information about the project, not concerning the data. For example a PowerPoint presentation on the experimental design of a study, or a contract or something else. Data information goes in the “supporting’ folder that is in the same folder as where the data file it refers to is stored.
Exercise 3
Provide study documentation and meta data for your last laboratory project.
While you are at it, add /code folders for any scripts you write (outside of the RMarkdown files) and /R folders for any functions you write.
3.7 What your data should look like
Now that we have a framework with which we can build our work flows in a data science project we can start working and collaborating. Below I resume some key concepts that are useful when working in a data science team.
3.7.1 Data formatting
This information is not new, but DAUR1 is a while ago, so we’ll repeat it: data can be formatted in different ways.
During the different R courses we have been working with data in the tidy format frequently.

- Each variable goes in its own column
- Each observation goes in its own row
- Each cell contains only one value
From: “R for Data Science”, Grolemund and Wickham
Although this is a optimized format for working with the {tidyverse} tools it is not the only suitable data format. We already encountered an important other structure that is much used in Bioinformatics: SummarizedExperiment. This class of data format is optimized for working with Bioconductor packages and work flows.

Morgan M, Obenchain V, Hester J, Pagès H (2021). SummarizedExperiment: SummarizedExperiment container. R package version 1.22.0, https://bioconductor.org/packages/SummarizedExperiment.
For machine learning purposes, data is often formatted in the wide format. We see an example here:
## # A tibble: 506 × 14
## crim zn indus chas nox rm age dis rad tax ptratio b
## <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.00632 18 2.31 0 0.538 6.58 65.2 4.09 1 296 15.3 397.
## 2 0.0273 0 7.07 0 0.469 6.42 78.9 4.97 2 242 17.8 397.
## 3 0.0273 0 7.07 0 0.469 7.18 61.1 4.97 2 242 17.8 393.
## 4 0.0324 0 2.18 0 0.458 7.00 45.8 6.06 3 222 18.7 395.
## 5 0.0690 0 2.18 0 0.458 7.15 54.2 6.06 3 222 18.7 397.
## 6 0.0298 0 2.18 0 0.458 6.43 58.7 6.06 3 222 18.7 394.
## 7 0.0883 12.5 7.87 0 0.524 6.01 66.6 5.56 5 311 15.2 396.
## 8 0.145 12.5 7.87 0 0.524 6.17 96.1 5.95 5 311 15.2 397.
## 9 0.211 12.5 7.87 0 0.524 5.63 100 6.08 5 311 15.2 387.
## 10 0.170 12.5 7.87 0 0.524 6.00 85.9 6.59 5 311 15.2 387.
## # ℹ 496 more rows
## # ℹ 2 more variables: lstat <dbl>, medv <dbl>The different variables are arranged in a side-by-side fashion. In this example the data is still tidy, but there are also examples of wide formatted data that is not tidy. When you want to work with this data, you generally need to transform it in a stacked or so-called long format that works well for {tidyverse}. We will see an example in the next exercise
Exercise 3
Transforming data in a reproducible way
Remember the well plate experiment? Here we will perform a transformation on the data to make the data suitable for analysis in R. We will also create a single graph showing all the data available for all samples over measured time in the experiment.
The data file for this exercise can be found here: ./data/salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx
Download it here if you’ve lost it and take a good look at your file management…
Go over the following steps to complete this exercise.
- Review your answers to the previous exercise where we used this file in this lesson.
- Try reading the sheet called
All Cyclesin the Excel file. - What goes wrong with the formatting of the data if you start reading in the data from cell A1?
- Try solving this problem.
- What shape of formatting would you say this data is in? Is the data tidy?
- Write a piece of code that creates a tidy format of this data. You also need to take a look a the sheet called
layoutto get information on the samples. Try generating a manual data frame that has 96 rows and a code for each sample. The experiment has been performed in duplo, so for each experimental condition there are two samples. - Now join your
sample datadataframe to the raw data. - Export the data as a .csv file.
- Write an appropriate README.txt file that accompanies this exported csv file. Save both in your
datafolder of your course project. (make this folder if you don’t have it yet).
**TIPS:
- Remember:
dplyr::pivot_longer()anddplyr::pivot_wider()are very helpful when you want to reshape your data in R - After reading your data into R: be sure to check the datatype of the columns
- Create a sample data table containing sample information for each of the 96 samples mentioned in the
All Cyclessheet. The information you need to do this is contained in sheetlayout - the
timevariable in this dataset is a nasty one. It is recorded in an uncenventional way. You need to use some cleaning up code to transform this variable in numbers (usestr_replace_all(), to get rid of the stupid characters likexand the_. Next you can transform this variable into hours and minutes by usingseperate(). Be aware that there will be ‘empty’ cells in theminutescolumn - Do you think it is a good idea to have graphs in an Excel worksheet that contains the raw data?
- How would you have stored the Raw data of this experiment?
Click for the answer
answer
# reading in the data - without any special settings
library(readxl)
data_platereader <- read_xlsx(
here::here(
"data",
"salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx"
), sheet = "All Cycles"
)
## this data looks mangled because of several things:
# there is some metadata in the top region of the sheet
# there is a weird looking headers (two headers?)
## trying skip
data_platereader <- read_xlsx(
here::here(
"data",
"salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx"
), sheet = "All Cycles", skip = 11
)
## clean up and fix names
data_platereader <- data_platereader %>%
rename(sample = Time, well = ...1) %>%
janitor::clean_names()
## which wells have data?
unique(data_platereader$well)## [1] "A01" "A02" "A03" "A04" "A05" "A06" "A07" "A08" "A09" "A10" "A11" "A12"
## [13] "B01" "B02" "B03" "B04" "B05" "B06" "B07" "B08" "B09" "B10" "B11" "B12"
## [25] "C01" "C02" "C03" "C04" "C05" "C06" "C07" "C08" "C09" "C10" "C11" "C12"
## [37] "D01" "D02" "D03" "D04" "D05" "D06" "D07" "D08" "D09" "D10" "D11" "D12"
## [49] "E01" "E02" "E03" "E04" "E05" "E06" "E07" "E08" "E09" "E10" "E11" "E12"
## [61] "F01" "F02" "F03" "F04" "F05" "F06" "F07" "F08" "F09" "F10" "F11" "F12"
## [73] "G01" "G02" "G03" "G04" "G05" "G06" "G07" "G08" "G09" "G10" "G11" "G12"
## [85] "H01" "H02" "H03" "H04" "H05" "H06" "H07" "H08" "H09" "H10" "H11" "H12"## create sample table
sample_names <- data_platereader$sample
mv_utr_tx100 <- rep(c("mv", "mv", "mv", "mv",
"untr", "untr", "untr", "untr", "untr",
"tx100", "tx100", "tx100"), times = 8)
salmonella <- read_xlsx(
here::here(
"data",
"salmonella CFU kinetics OD600 in LB van ipecs 8okt2020 kleur.xlsx"
), sheet = "layout", range = "C5:N13"
) %>%
janitor::clean_names()
# cheack data types
map(
.x = salmonella,
typeof
)## $ul_sal_1
## [1] "double"
##
## $ul_sal_2
## [1] "double"
##
## $ul_sal_3
## [1] "double"
##
## $ul_sal_4
## [1] "double"
##
## $ul_sal_5
## [1] "double"
##
## $ul_sal_6
## [1] "double"
##
## $ul_sal_7
## [1] "double"
##
## $ul_sal_8
## [1] "double"
##
## $ul_sal_9
## [1] "double"
##
## $ul_sal_10
## [1] "double"
##
## $ul_sal_11
## [1] "double"
##
## $ul_sal_12
## [1] "double"salmonella <- salmonella %>%
pivot_longer(ul_sal_1:ul_sal_12,
names_to = "plate_column",
values_to = "microliters_bacteria")
## synthesize to sample table
samples <- tibble(
well = data_platereader$well,
sample = sample_names,
condition = mv_utr_tx100,
ul_salmonella = salmonella$microliters_bacteria
)
## join sample table with data
data_join <- left_join(samples, data_platereader)
## create tidy version
data_tidy <- data_join %>%
pivot_longer(
x0_h:x24_h_5_min,
names_to = "time",
values_to = "value"
)
## fix time variable
data_tidy_time <- data_tidy %>%
mutate(time_var =
str_replace_all(
string = time,
pattern = "x",
replacement = ""
)) %>%
mutate(time_var =
str_replace_all(
string = time_var,
pattern = "_",
replacement = ""
)) %>%
mutate(time_var =
str_replace_all(
string = time_var,
pattern = "h",
replacement = ":"
)) %>%
mutate(time_var =
str_replace_all(
string = time_var,
pattern = "min",
replacement = ""
)) %>%
separate(
col = time_var,
into = c("hours", "minutes"),
remove = FALSE
) %>%
mutate(
minutes = ifelse(minutes == "", "0", minutes)
) %>%
mutate(minutes_passed = 60*as.numeric(hours) + as.numeric(minutes))
## misingness
data_tidy %>%
naniar::vis_miss()
## graphs
data_tidy_time %>%
group_by(condition, ul_salmonella, minutes_passed) %>%
summarise(mean_value = mean(value)) %>%
mutate(ul_salmonella = round(as.numeric(ul_salmonella), 2)) %>%
ggplot(aes(x = minutes_passed, y = mean_value)) +
geom_line(aes(colour = condition), show.legend = FALSE) +
facet_grid(condition ~ ul_salmonella) +
xlab("Time passed (minutes)") +
ylab("Mean AU")
3.7.2 Variable encodings
- Use explicit encoding: male/female instead of
0/1 - Encodings can always be altered programmatically
- Be consistent (see Figure 3.2)
- Write documentation that explains encodings, including units and levels
- Use factors if a variable has a set of discrete possible outcomes:
sex,species,marital_statusetc - Use an ordered factor if there is a hiearchy in the factor levels: e.g.
year,month, etc
Here we use the {palmerpenguins} dataset as an example to show you how they dealt with encoding variables.
# install.packages("remotes")
# remotes::install_github("allisonhorst/palmerpenguins")
library(palmerpenguins)
data_penguins <- palmerpenguins::penguins_raw
data_penguins## # A tibble: 344 × 17
## studyName `Sample Number` Species Region Island Stage `Individual ID`
## <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
## 1 PAL0708 1 Adelie Penguin… Anvers Torge… Adul… N1A1
## 2 PAL0708 2 Adelie Penguin… Anvers Torge… Adul… N1A2
## 3 PAL0708 3 Adelie Penguin… Anvers Torge… Adul… N2A1
## 4 PAL0708 4 Adelie Penguin… Anvers Torge… Adul… N2A2
## 5 PAL0708 5 Adelie Penguin… Anvers Torge… Adul… N3A1
## 6 PAL0708 6 Adelie Penguin… Anvers Torge… Adul… N3A2
## 7 PAL0708 7 Adelie Penguin… Anvers Torge… Adul… N4A1
## 8 PAL0708 8 Adelie Penguin… Anvers Torge… Adul… N4A2
## 9 PAL0708 9 Adelie Penguin… Anvers Torge… Adul… N5A1
## 10 PAL0708 10 Adelie Penguin… Anvers Torge… Adul… N5A2
## # ℹ 334 more rows
## # ℹ 10 more variables: `Clutch Completion` <chr>, `Date Egg` <date>,
## # `Culmen Length (mm)` <dbl>, `Culmen Depth (mm)` <dbl>,
## # `Flipper Length (mm)` <dbl>, `Body Mass (g)` <dbl>, Sex <chr>,
## # `Delta 15 N (o/oo)` <dbl>, `Delta 13 C (o/oo)` <dbl>, Comments <chr>Make sure you are consistent in entering the data!
library(ggplot2)
# simulating inconsistent data entry
penguinswrong <- penguins
levels(penguinswrong$species) <- c(levels(penguinswrong$species), "adelie")
penguinswrong$species[1:5]<-"adelie"
# make a box plot of flipper length showing a/Adelie as separate species
flipper_box <- ggplot(data = penguinswrong, aes(x = species, y = flipper_length_mm)) +
geom_boxplot(aes(color = species), width = 0.3, show.legend = FALSE) +
geom_jitter(aes(color = species), alpha = 0.5, show.legend = FALSE, position = position_jitter(width = 0.2, seed = 0)) +
scale_color_manual(values = c("darkorange","purple","cyan4","red")) +
theme_minimal() +
labs(x = "Species",
y = "Flipper length (mm)")
flipper_box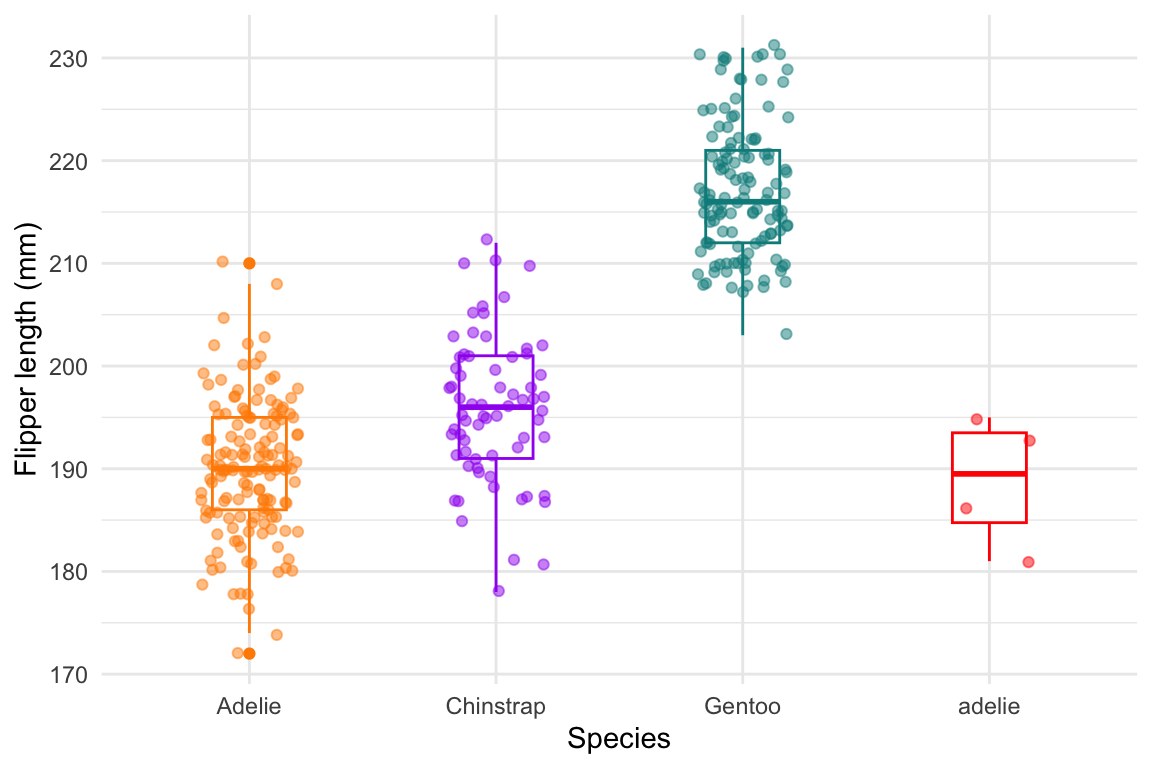
Figure 3.2: Example of inconsistent data entry
3.7.3 Factor levels
R (unlike SPSS) does not mind if you use descriptive words instead of numbers as categorical variable values. This increases reproducility! GGplot doesn’t mind either. (Machine learning workflows may mind, but we’re not doing machine learning here.) The different possible options for such a variable are called the levels of this factor:
data_penguins %>%
ggplot(aes(x = Sex, y = `Flipper Length (mm)`)) +
geom_point(aes(colour = Species), position = "jitter", show.legend = FALSE)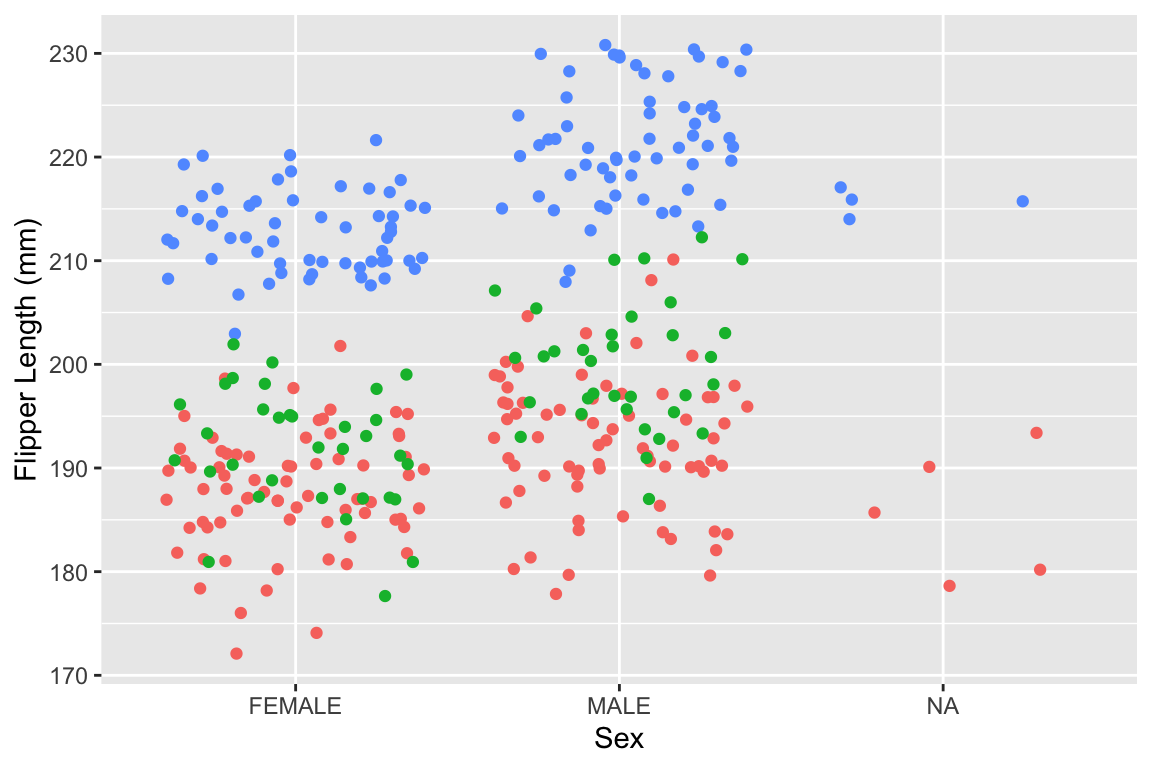
Figure 3.3: Graph displaying the flipper length per factor level of Sex
## [1] "MALE" "FEMALE" NA3.7.4 Data-formats - Non-Proprietary
When we store data for re-use, we need it to be in an interoperable form. This means that it can be read (also after a long time - let’s say 30 years from now) into analysis software. This can be achieved by storing data in a so-called non-proprietary format. This means basically that the format source code is open and maintained by open source community or core development team.
Here are some examples:
.netCDF(Geo, proteomics, array-oriented scientific data)
.xml/.mzXML(Markup language, human and machine readable, metadata + data together).txt/.csv(flat text file, usually tab, comma or semi colon (;) seperated).json(text format that is completely language independent)fastq/fastaand their equivalents
These formats will remain readable, even if the format itself becomes obsolete
When storing a curated dataset for sharing or archiving it is always better, and sometimes enforced by the repository, to choose a non-proprietary format
3.7.5 Data entry
Data entry preferably, must be performed in a project template. The template contains predefined information on the observations in the study. The blank information needs to be filled out by the person responsible for/or performing the data entry. Or in other words: think about how you will enter your data before gathering it, and if there are multiple people gathering the data, make sure that everyone uses the exact same way of entering the data (template).
Enter an “NA” for missing values, do not leave cells blank if there is a missing value. Use only “NA” and nothing else. If you want to add additional information on the “NA”, put that in the “remarks” column.
(By the way, you can visualise missing data like this in R with the naniar package: )

Or check out a ggplot method here.
After entry (and validation) of the filled-out template, NEVER change a value in the data. If you want to make changes, increment the version number of the file and document the change in the README.txt file or sheet in an Excel file (see below)
3.7.6 Tidy template
A tidy data template you may want to use is available here
When you are planning to use this template, please be aware the following pointers:
For Excel users:
- Start your file in A1 of a clean new sheet.
- Use row 1 for variable names, use the rest of the rows for observations
- It is allowed to have existing sheets in the file. Label the new tidy formatted datasheet as “tidy”
- Do not fuse cells
- Provide one value per cell so e.g. put units in a separate column
- Adhere to the naming conventions for variable names (see above)
- Use cell validation for entered values. This is especially true if you have multiple categorical variables that need manual labeling in Excel. A typo is just around the corner.
Portfolio assignment
Maak opdracht 3a en 3b van de portfolio-opdrachten.
3.8 Resources
- F1000 ‘Data guidelines’
- Guerrilla Analytics
- Guerrilla Analytics ‘slides’
- F1000 ‘Preparing your data for sharing’
CC BY-NC-SA 4.0 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. Unless it was borrowed (there will be a link), in which case, please use their license.
